Using the Sequence Data module, one can assemble sequence reads ranging from single genes obtained via classical Sanger sequencing to full genomes via Next Generation Sequencing (NGS). In addition, the BIONUMERICS software recognizes widely used sequence file formats such as EMBL, GenBank, and FASTA, and stores the full header and feature information of annotated sequences.
With the addition of the Sequence Data module, BIONUMERICS becomes a rich and uniquely integrated sequence analysis software, offering a variety of analysis tools such as multiple sequence alignment, phylogenetic clustering, chromosome comparison and alignment, automated annotation, chromosome-wide SNP analysis, PCR primer design, restriction enzyme analysis, open reading frame analysis, etc.
BIONUMERICS' powerful database back end supports sequence analysis in high throughput projects. Furthermore, available plugins, script tools and pipeline engines allow for reliable automated batch processing of massive amounts of sequence data.
Manage next generation sequencing data
The sequence read sets experiment type offers an integrated environment for importing, preprocessing and analyzing sets of reads from high throughput sequencers or public repositories.
Multiple sequence alignment
BIONUMERICS offers probably the finest and most comprehensive multiple sequence alignment tool that currently exist for PCs. It combines clustering of thousands of nucleotide or protein sequences of almost unlimited length with multiple alignment and display of homology matrices.
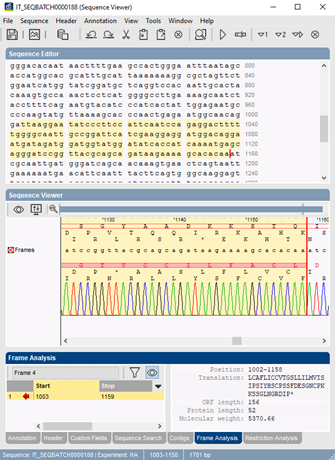
Open reading frame analysis
Frame analysis finds all open reading frames (ORF) and predicts protein coding sequences (PCS) on a sequence. The open reading frame finder in BIONUMERICS lists all ORFs and possible PCS regions for all six translation frames, for a given translation table and codon usage table. The user can specify an optional ORF and PCS length filter. ORFs and amino acid translations can be plotted on a graphical sequence display.
Primer design
The PCR primer design tool in the BIONUMERICS software searches for optimal primers or primer combinations for the most diverse experiment setups by taking into account a large number of experimental parameters. The user can specify various primer properties such as preferential length and melting temperature, %GC boundaries, maximal degeneracy, etc. Forward and reverse primers and PCR combinations can be sorted and selected according to different parameters (position, melting temperature, length, degeneracy, %GC...). Selected primer pairs can be stored in the oligonucleotide database.
Restriction enzyme analysis
In-silico multi-purpose analysis of restriction enzyme cleavage suitable for cloning experiments as well as for RFLP, PFGE and AFLP design. Thousands of restriction enzymes from ReBase can be downloaded and stored, and subsets of enzymes of particular interest can be created (e.g. 4-cutters, blunt-cutters, cheap enzymes, available enzymes...). Up to full chromosomes can be analyzed so that optimal enzymes can be selected for electrophoresis-based fragment typing techniques.
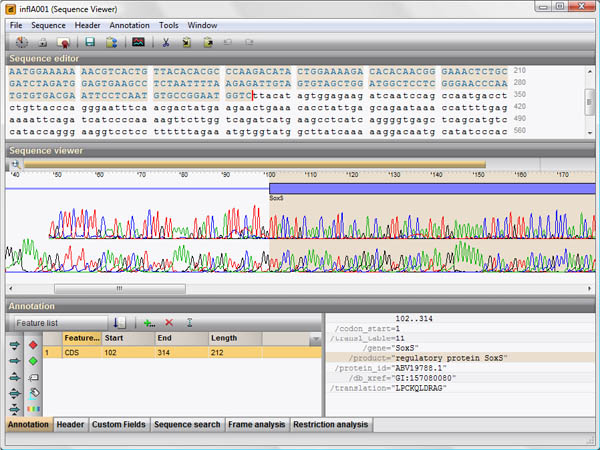
Sequence assembly from trace files
BIONUMERICS’ sequence assembly tool allows direct import of raw trace files from Sanger sequencing, i.e. generated by an ABI, Beckman or MegaBace automated sequencer. The assembly software combines a powerful alignment engine with an informative and intuitive interface.
Sequence import from GenBank, FASTA and other text formats
Direct import of of nucleic acid and amino acid sequences from text files in EMBL, GenBank, Flat A, or FASTA format into a BIONUMERICS database. Easy copy & paste from clipboard, and manual sequence editing tools.
SNP analysis
BIONUMERICS’ multiple sequence alignment tool is an invaluable asset for single nucleotide polymorphism (SNP) and mutation analysis. SNPs or mutations are screened for through up to many thousands of aligned sequences.
