BIONUMERICS lets you align and compare sequences of up to full chromosome length. With these comparative genomics tools, discontinuous alignments are calculated using seed and stretch-based sequence mapping, revealing genomic inversions, swaps, duplications, insertions and deletions. Mutation and SNP discovery can be performed on template-based multiple chromosome alignments, with optional selection of mutation type (intergenic, synonymous, non-synonymous or indel) and filtering of significant SNPs based on quality scores. dNdS analysis based on the ratio synonymous/non-synonymous mutations within gene clusters is available to predict evolutionary selection pressure on genes.
N×N pairwise chromosome comparison
- Extremely fast pairwise matching and alignment of N×N sequences of full chromosome size.
- Seed and stretch based sequence matching, allowing for discontinuous alignments, including inversions, swaps, duplications, insertions and deletions.
- Full-sequence based or coding sequence based alignment, using user-defined nucleic acid seeds and/or amino acid seeds.
- Display of clickable N×N chromosome similarity matrix and associated clustering. The selected pair of chromosomes is displayed in the dot plot Cell panel. Direct and inverted matches are displayed in different color.
- Clickable dot plot matrix, updating (1) currently selected stretch in the Stretches panel and (2) associated alignment in the Pairwise alignment panel.
- Calculation of superstreches, i.e. larger clusters of stretches that contain no major discontinuities and that have a minimal overall homology.
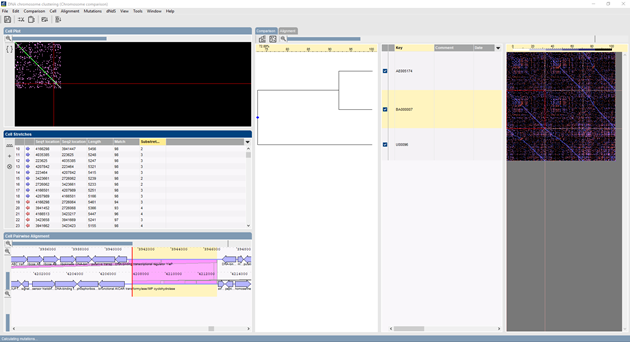
Template-based multiple chromosome alignment
- Alignment of multiple chromosomes based upon selected template chromosome.
- Display of overview panel and detailed multiple alignment panel.
- Synchronized selection in overview, multiple alignment, dot plot, stretches and pairwise alignment panels.
- Versatile search and display functions for features and subsequences on multiple chromosomes.
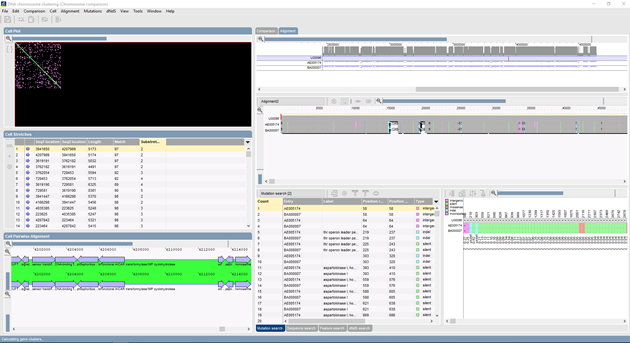
Mutation and SNP analysis
- Search and display of mutations within multiple alignments, with discrimination between intergenic, synonymous, non-synonymous and Indel mutations.
- Additional filtering based on SNP quality scores.
- Display colors based on mutation type or quality; sorting based on position, gene, NA change, AA change, quality (see figure above, Mutation analysis panel).
- Direct clustering based upon mutations or export of mutation list for further analysis.
dNdS ratio analysis
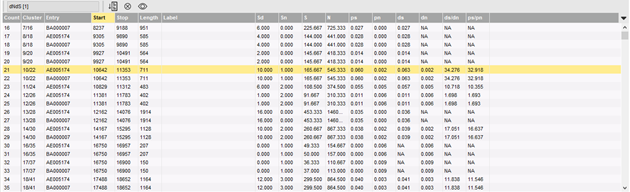
The dNdS ratio for a gene compares the non-synonymous mutations versus the synonymous mutations and as such, provides information on the positive selection pressure of the gene. The starting point in BIONUMERICS is a template-based multiple chromosome alignment. For each ORF on the template sequence, the aligned ORFs on the other sequences (if any) are grouped and synonymous/non-synonymous mutations are calculated for each codon. A list of dNdS ratios and related parameters for all ORFs is calculated, and the dNdS ratios can optionally be plotted in differential color on the multiple alignment.