ANOVA and MANOVA
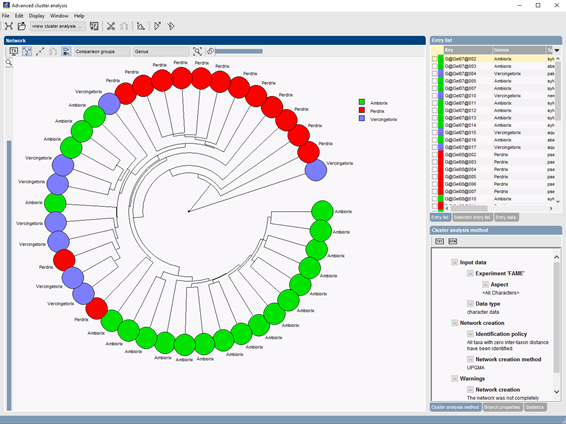
BIONUMERICS offers a generalized and well-documented implementation of ANOVA (Analysis of Variance) and MANOVA (Multivariate Analysis of Variance) with comprehensive statistical analysis and validation testing tools. These very useful statistical methods allow you to investigate the relation between groups of entries and characters, as well as the significance of such groups. The groups can be clusters derived from a dendrogram, or any user-defined selections of entries (e.g., by origin, species, serotype …).