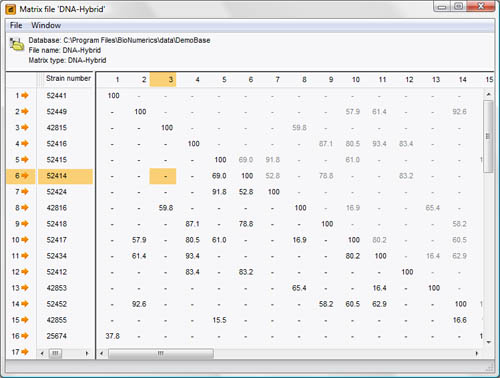
An externally generated distance matrix or similarity matrix can be imported and linked to database entries in a BIONUMERICS database. This is used in conjunction with other information to obtain classifications and identifications. The distance or similarity values are either measured directly by the technique (a typical example being DNA-DNA hybridization values in bacterial taxonomy), or generated by other software.
Specifications:
Easy import of similarity or distance matrices. Similarity matrices can also be generated or imported using coefficients designed by the user. Partial matrices (e.g. DNA homology matrices) are supported: BIONUMERICS contains a special UPGMA-based clustering algorithm for incomplete matrices.
Supported in BIONUMERICS configurations:
BIONUMERICS-GEL
BIONUMERICS-MALDI
BIONUMERICS-SEQ
BIONUMERICS-SUITE
